
- #Clamxav database mirror install
- #Clamxav database mirror update
- #Clamxav database mirror Patch
- #Clamxav database mirror verification
- #Clamxav database mirror code
Next, you'll need to configure the freshclam clients so they'll update from your private mirror.įor nf on your downstream freshclam clients, set: # Replace `mirror.mylan` and `8000` with your domain and port number. Once you have the database files, host them with your favorite webserver, or use the cvd serve test-webserver (not intended for production). For ore detailed instructions, or to report issues, please visit: ]() Tip: You can use -help with any cvd command to learn more. If you didn't set a custom database path, the databases will be stored in ~/.cvdupdate/database You may wish to set up a cron job to check for updates. Now run this as often as you need, or at least once a day to download/update the databases: cvd update (optional) Once installed, you may wish to configure where the databases are stored: cvd config set -dbdir
#Clamxav database mirror install
You can easily install cvdupdate using Python 3's Pip package manager: pip3 install cvdupdate cvdupdate, on the otherhand, will download both the new daily CDIFF and the daily CVD every day. freshclam is far more efficient, even for a small cluster of installs, because it will update with CDIFF patches after the initial database downloads. IMPORTANT: Please do NOT use cvdupdate if you don't need to host a private database mirror. It should work fine on Linux/Unix and on Windows.An internet connection with DNS enabled.These instructions use a tool named cvdupdate.
#Clamxav database mirror Patch
This means that your downstream freshclam clients will be able to update using the CDIFF patch files, which should save some bandwidth between your private mirror and your clients. This solution will allow you to host a mirror that functions in the same way as the official database CDN, serving CVD and CDIFF files. You may use a tool named cvdupdate on a private mirror to maintain the latest CVD databases and CDIFF patch files. Use cvdupdate to serve whole databases and database patch files from a private mirror
Sometimes the servers which perform the scan are not directly connected to Internet and can only download updates from a server in the same network segment.įor people who face these problems, we recommend using one of the following solutions: This is a waste of bandwidth and resources for your network and for our mirrors network. If you run ClamAV on many clients on your network, each new installation will download a copy of the database files. There are some situations in which it may be desirable to set up a private mirror for distributing ClamAV databases.
#Clamxav database mirror verification
Microsoft Authenticode Signature Verification Selecting the Right Version of ClamAV for You * HF (high fidelity) versions of these enzymes are available.
#Clamxav database mirror code
| A | B | C | D | E | H | K | M | N | P | R | S | T | X | single letter code EnzymeĪciI, AclI, BsaHI (GR/CGYC), HinP1I, HpaII, NarIĪccI (GT/CGAC), AclI, ClaI, BstBI, TaqI-v2ĪccI (GT/CGAC), AciI, ClaI, BstBI, HinP1I, HpaII, NarI, TaqI-v2ĪpaI, BanII, Bsp120I, Bsp1286I, HaeIII, NlaIV, Sau96IĪgeI, BsaWI, BspEI, BsrFI (R/CCGGY), NgoMIV, SgrAI (CR/CCGGYG)īanI, BsaHI, HaeII, HhaI, KasI, NarI, NlaIVĪpaI, BanII, Bsp1286I, HaeIII, NlaIV, Sau96IĪgeI, BsaWI, BsrFI (R/CCGGY), SgrAI (CR/CCGGYG)īsiHKAI (GTGCA/C), Bsp1286I (GTGCA/C), PstI, SbfIīanII (GAGCT/C), BsiHKAI, Bsp1286I (GAGCT/C)ĪvaI, BsaJI, HpaII, NciI, ScrFI, SmaI, XmaI
Be aware that these degenerate enzymes will cleave sequences in addition to the one listed.Ī "-" denotes a ligation product that cannot be recleaved. recognize more than one sequence) are followed by a specific sequence in parentheses and are only listed if a non-degenerate equivalent does not exist. Only enzymes available from New England Biolabs have been listed.Įnzymes that have degenerate recognition sequences (i.e. Where isoschizomers exist, only one member of each set is listed. The combinations listed were identified by computer analysis, and although we have tried to ensure their accuracy, they have not necessarily been confirmed by experimentation. Restriction endonucleases that produce compatible cohesive ends often produce recleavable ligation products. Cleavage with two restriction endonucleases that produce compatible overhangs.Cleavage with two restriction endonucleases that produce blunt ends.Cleavage followed by fill-in of 5´ overhangs to generate blunt ends.These new restriction sites may be generated by: New restriction sites can be generated by ligation of DNA fragments with compatible cohesive or blunt ends. Home Tools & Resources Selection Charts Compatible Cohesive Ends and Generation of New Restriction Sites Compatible Cohesive Ends and Generation of New Restriction Sites


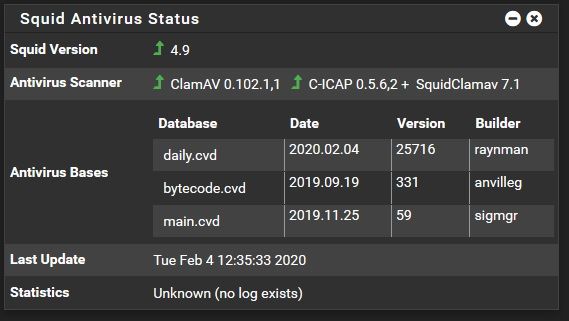
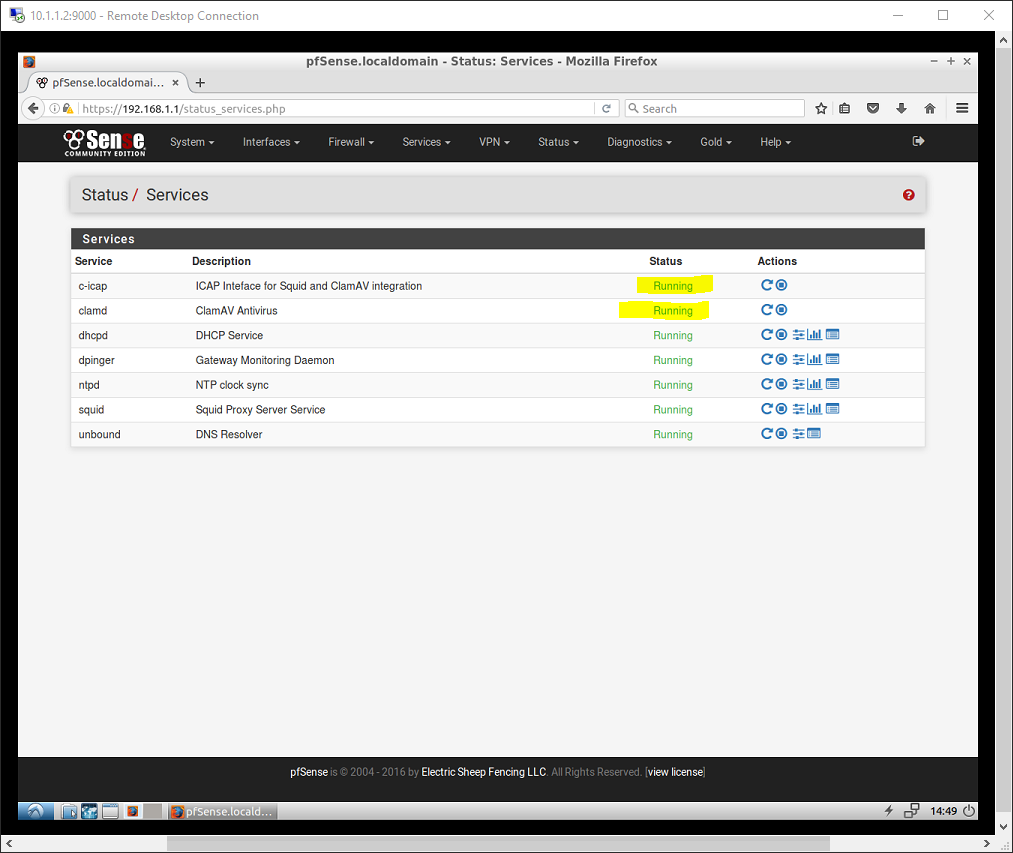
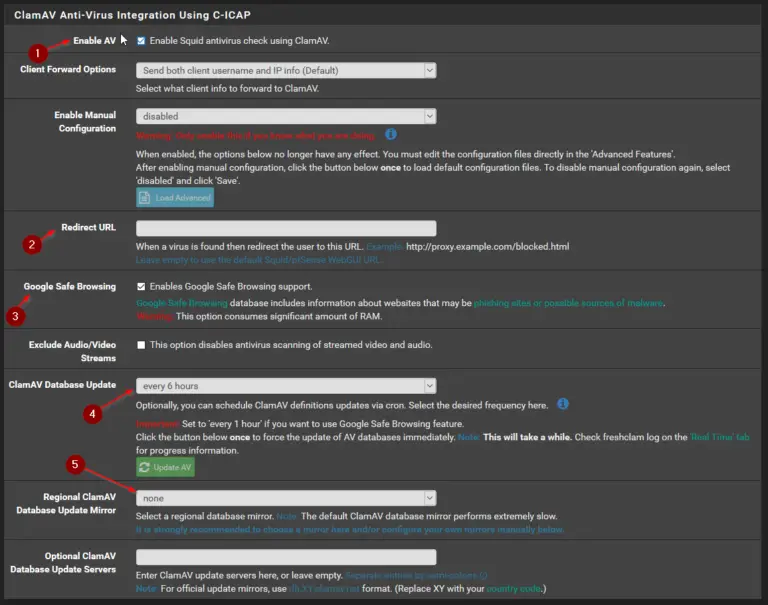
 0 kommentar(er)
0 kommentar(er)
